Disease networks from ML-driven mining of curated knowledge
 Example disease network
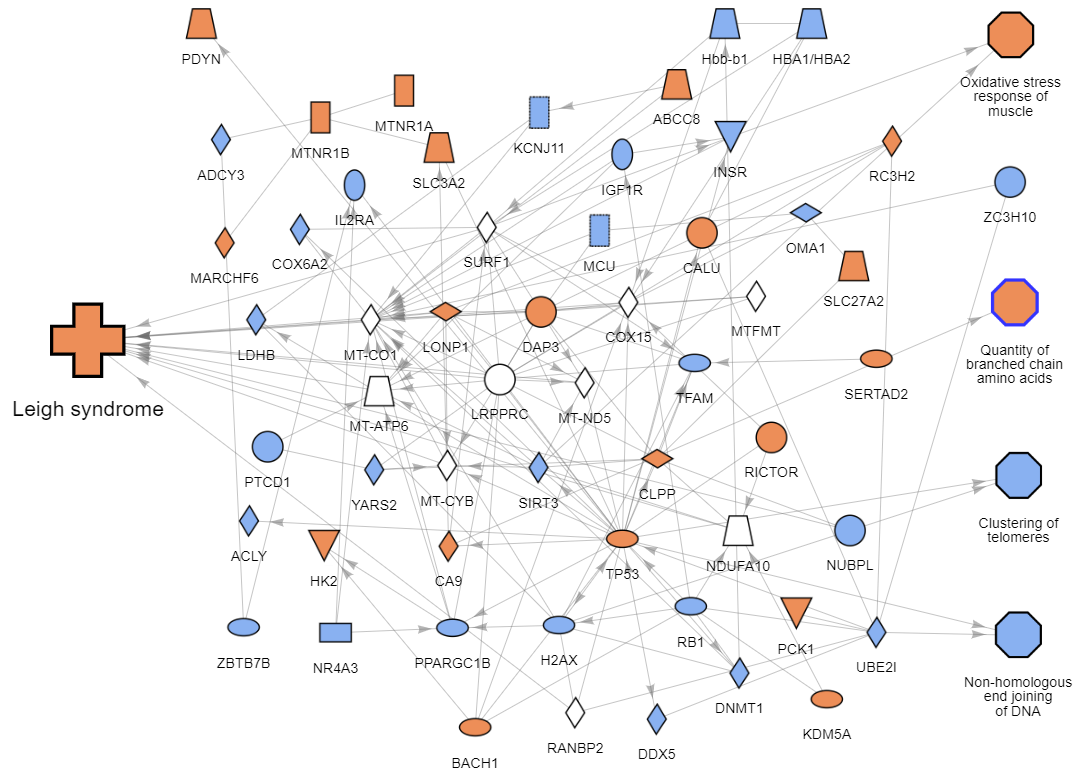
Example disease network
Each network in the collection focuses on a single disease or phenotype and contains key genes, impacted biological functions, and relationships between them that drive the condition. In addition, a colored pattern of predicted activation is overlaid to show how the activation (orange) or inhibition (blue) of genes leads to the disease.
The intent is to show a relatively small snapshot of the primary factors involved. The network does not contain all molecules known to be related to the disease in the QKG; including all would often result in an unreadable, densely connected network with hundreds, if not thousands, of nodes. Instead, the ML algorithm prioritizes the most important genes and functions and generates networks of reasonable size (~50 nodes on average) that provide a good overview in a comprehensible manner.
To make such prioritizations, the algorithm utilizes unsupervised gene and function embeddings derived from causal relationships in the QKB. Unlike many ML applications for biology, the algorithm does not train on differential expression or other forms of raw data; instead, it leverages the QKB's causal associations curated from biomedical literature by experts over more than 20+ years. More details about the approach are available in our recently published paper (1).
The results are generated algorithmically without further curation by human experts. Each network generally includes well-known participants in the disease and also predicts many new associations not previously present in the QKB. Some of these predictions may be opportunities for novel discoveries, and in fact several have already been confirmed by manually searching across literature not yet curated for the QKB.
For example, the gene FGF21 appears in the network "Lung squamous cell carcinoma." In the QKB, this gene is not yet associated with the disease, but the ML algorithm predicts a significant activating relationship and, therefore, includes it in the network. As it turns out, recently published research suggests the gene is a promoter of lung cancer and a potential therapeutic target (2). Similarly, the "Leigh syndrome" network predicts that the mitochondrial tyrosyl-tRNA synthetase YARS2 is inhibited when the disease occurs, and a recent study linked mutated YARS2 (i.e. loss of activity) to ocular dysfunction, one of the disease's clinical features (3).
Disease networks like these, generated by applying ML techniques to the QKB, contain both expected and novel genes and are therefore, a useful tool for understanding the drivers of a disease as well as discovering potential new players. Over 1500 such networks are currently available in Ingenuity Pathway Analysis (IPA) as a preview, and several are also publicly available here in this Disease Network Explorer.
1. Mining hidden knowledge: Embedding models of cause-effect relationships curated from the biomedical literature. Krämer A, et al. (2022) Bioinformatics Advances. vbac022. https://doi.org/10.1093/bioadv/vbac022
2. FGF21 promotes non-small cell lung cancer progression by SIRT1/PI3K/AKT signaling. Yu X, et al. (2021) Life Sci. 269:118875. PMID: 33310036
3. An animal model for mitochondrial tyrosyl-tRNA synthetase deficiency reveals links between oxidative phosphorylation and retinal function. Jin X, et al. (2021) J Biol Chem. 296:100437 PMID: 33610547